A multiomic & phenotypic map of ageing as a foundational resource for the community.
Key features:
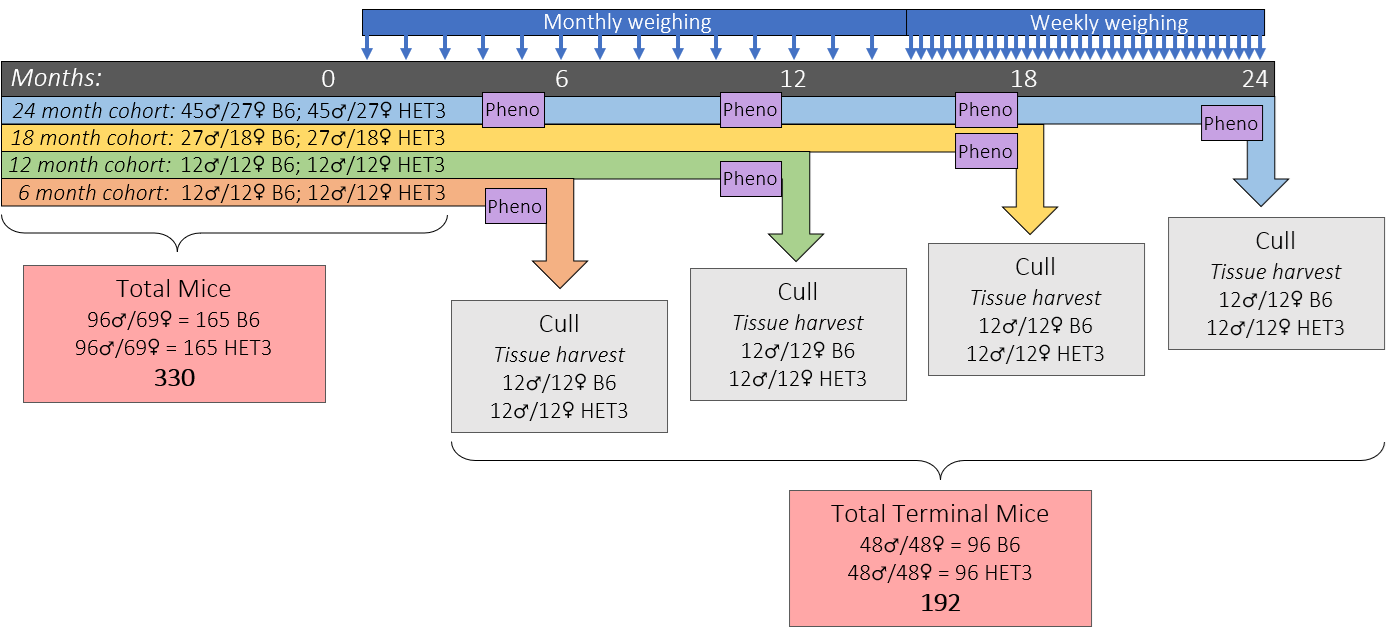
Two mouse strains
Inbred C57BL/6 and genetically diverse 4-way cross HET3
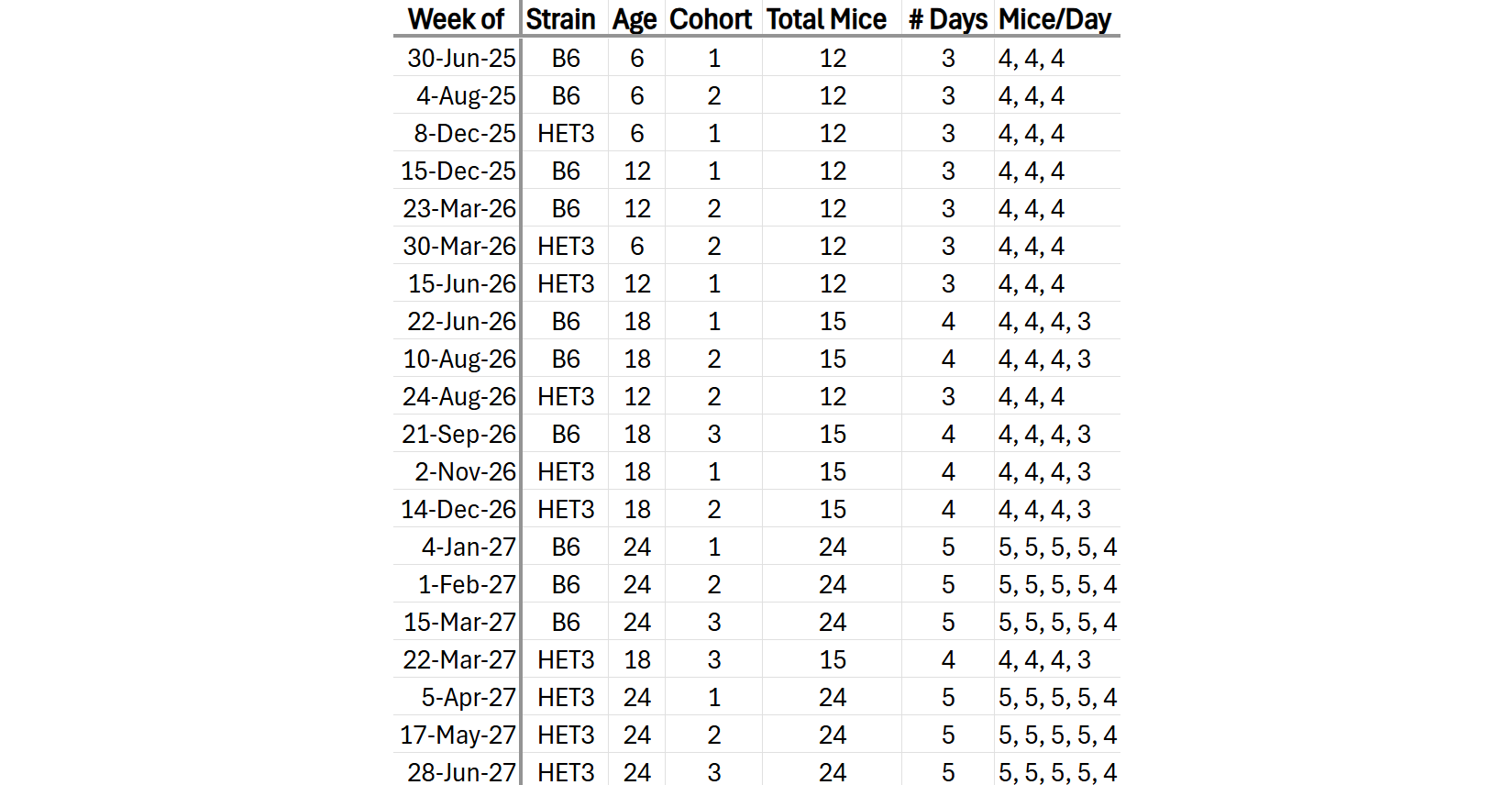
4 timepoints across the lifespan
6, 12, 18, 24mos
Longitudinal physiological phenotyping
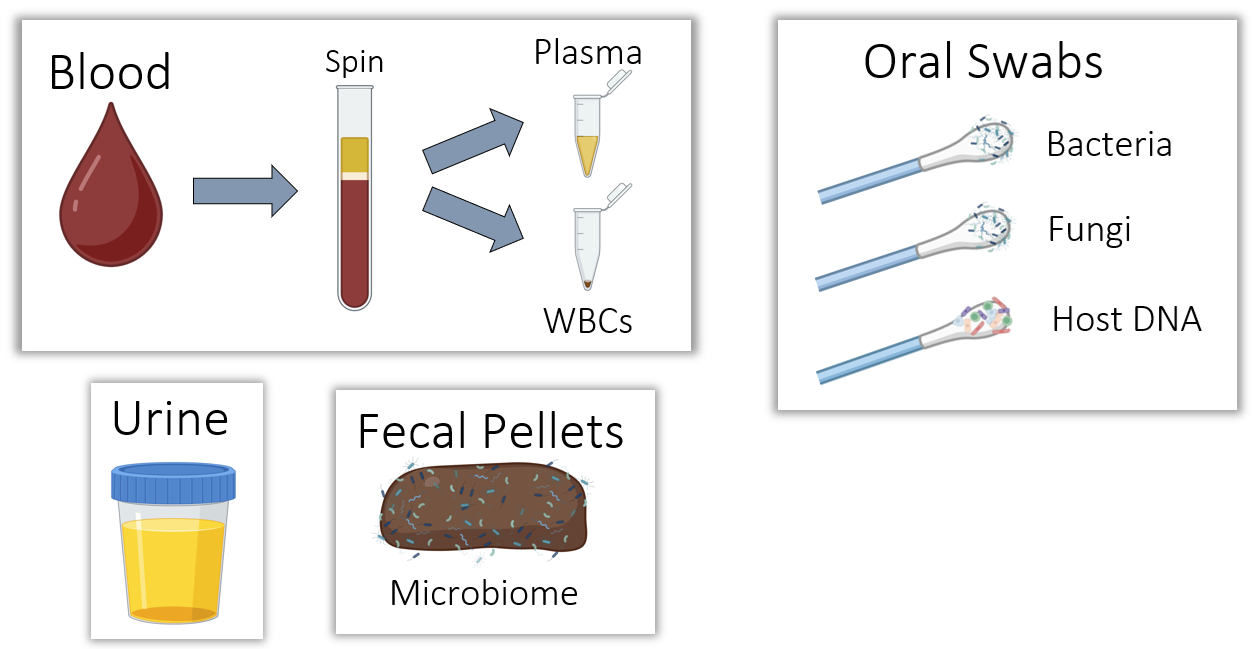
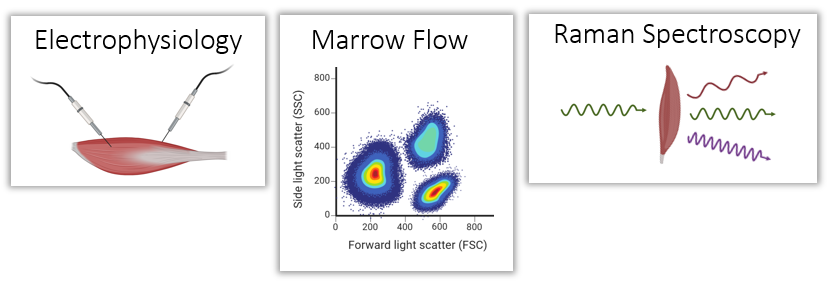
Multiomics of 4 tissues
Brain, muscle, intestine, immune
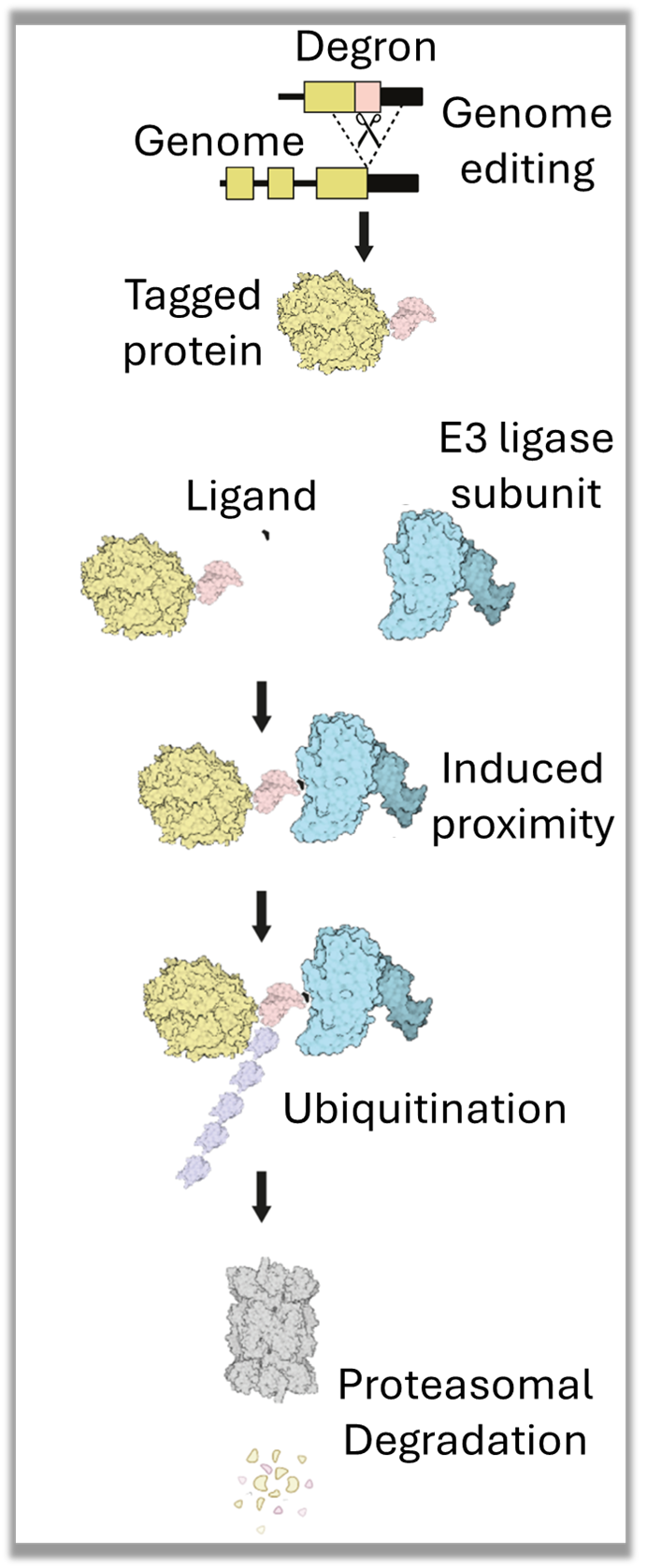
Development of 2 mouse models
Atg5-AID2 autophagy inhibition accelerated ageing
Cxcr4-AID2 vaccine response enhancement
Reach out to collaborate
〰️
Reach out to collaborate 〰️
Walid Khaled Cluster Lead Cambridge
Laura Greaves Cluster Lead Newcastle
Anne Ferguson-Smith Cambridge
Geula Hanin Ferguson-Smith Lab Cambridge
Michelle Linterman Babraham
Masashi Narita Cambridge
Dervis Salih UCL
Nick Schaum Khaled Lab Cambridge
Karen Suetterlin Newcastle
Helen Tuppen Greaves Lab Newcastle